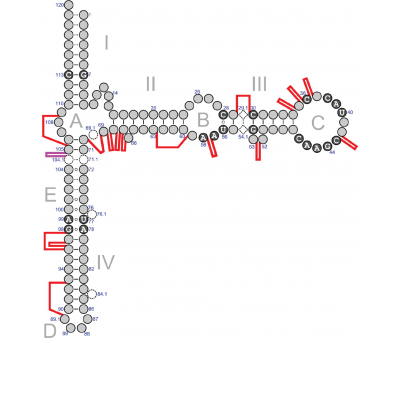
5S rRNA is an essential part of the central protuberance in the large ribosomal subunit. Although its exact function is not known, it is believed to be involved in intraribosomal interactions, which impose evolutionary constraints that make this small RNA one of the most conserved ribosomal components among all three domains of life. Thus, examining structural abnormalities such as expansion segments could be useful in studying the function of 5S rRNA and in ribosome redesign for biotechnological applications. However, the occurrence of oversized 5S rRNAs has not been well documented. This is likely because a considerable amount has been incorrectly presented as much shorter sequences in genomic annotations and databases to fit the consensus-derived standard model of 5S rRNA. The authors report that they screened the RFam RF00001 and 5SrRNAdb databases and the entire RefSeq collection of bacterial and archaeal genomes for expanded 5S rRNAs. With the discovery of 87 novel oversized 5S rRNAs, we now have a list of 89 specimens of 15 structural types among 15 archaeal and 36 bacterial strains. Each 5S rRNA molecule had 1-3 expansion segments ranging from 13 to 109 nts across 17 insertion sites. These insertions do not seem to interfere with the folding of the 5S rRNA core due to lack of complementarity and their extensive internal base pairing, which can form helices. But not all insertions appear compatible with the immediate surroundings of the 5S rRNA. In addition, genomes of some species contained very similar sequences to 5S rRNA genes and may be pseudogenes. Therefore, if only expanded 5S rRNA is present in the cell, it is assumed to be functional; but, if both normal and expanded 5S rRNA genes are present, it is uncertain whether the expanded 5S rRNA are active. In both archaea and bacteria, the expansion segments are observed in only a tiny fraction of 5S rRNAs and are mostly divided in taxonomically remote and structurally unrelated groups. Thus, even if they contribute to the organism’s survival, it is likely restricted to conditions uncommon for many prokaryotes. Analysis of prokaryotic environmental preferences shows a correlation between the presence of expanded 5S rRNA genes and microaerobic/anaerobic prokaryotes with strong halophilic or thermophilic traits. But it is also possible that some expansion segments are accidentally acquired through neutral evolution and do not provide adaptive advantages.
Stepanov, V. G., & Fox, G. E. (2021). Expansion segments in bacterial and archaeal 5S ribosomal RNAs. RNA (New York, N.Y.), 27(2), 133–150. https://doi.org/10.1261/rna.077123.120
