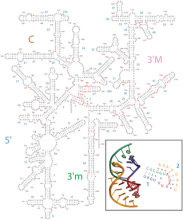
The structure of the ribosomal core is conserved in all living things, while the outer regions are highly variable. Accurate secondary structures are important for understanding ribosomes. Using 3D structures of ribosomes as input, we have corrected traditional secondary structures of rRNAs. We have generated secondary structures for both large subunit (LSU) 23S/28S and small subunit (SSU) 16S/18S rRNAs of Escherichia coli, Thermus thermophilus, Haloarcula marismortui (LSU rRNA only), Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens. Both LSU and SSU secondary maps are available (http://apollo.chemistry.gatech.edu/RibosomeGallery).
