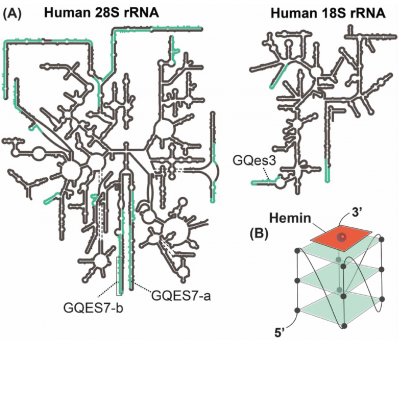
Heme is an essential signaling molecule in cells, but it must be tightly regulated due to its potential toxicity. Although the proteins that synthesize and degrade heme have been extensively studied, the regulation of heme bioavailability is currently poorly understood. The authors demonstrate that human rRNA tentacles form G-quadruplexes (G4s) in vivo that regulate cellular heme homeostasis. To study the potential formation of rRNA G4s in vivo, the authors performed confocal microscopy and RNA pulldown experiments. The confocal microscopy experiments suggested that a fraction of human ribosomes form G4s in vivo and that most extra-nuclear RNA G-quadruplexes are ribosomal, in agreement with the massive abundance of rRNA in cells with respect to other RNAs. Treating cells with PhenDC3, a G4 ligand known to induce and stabilize G4s, increased the ribosomal signal that colocalized with G4s, suggesting PhenDC3 promotes ribosomal G4 formation in vivo. Additionally, using BioTASQ (a small molecule that binds to G4-forming RNAs), human LSU rRNA and, to a lesser extent, SSU rRNA were pulled down from HEK293T cells. This agrees with the observation of more G-tracts in LSU rRNA than in SSU rRNA and strongly suggests that human ribosomes form G4s in vivo.
G4s have been reported to bind heme with high affinity. For this reason, the authors sought out to determine whether one of the potential physiological functions of rRNA G4s is to recruit heme to the ribosome. To this end, rRNA oligomers derived from G4 regions of human LSU and SSU rRNAs were designed and titrated into fixed amount of hemin, causing an increase in the hemin Soret band. Larger human ribosomal components purified from cells also induced changes in the hemin Soret band when in the presence of hemin, providing strong evidence of heme association with G4s of human ribosomes in vitro. To determine if these interactions occur in vivo, HEK293T lysates were incubated with with hemin-agarose. The results provide strong evidence that when cells are depleted of heme, a greater fraction of human LSU rRNA and, to a lesser extent, SSU rRNA bind to hemin-agarose than in heme replete conditions, indicating that human ribosomes bind endogenous heme in cells. To determine whether rRNA G4s are involved in this in vivo interaction, cells were treated with the G4 ligand PhenDC3. Under these conditions, an increased amount of LSU rRNA bound to hemin-agarose. A corresponding but weaker signal is seen for the SSU, which has fewer G4 regions. The authors provide a model supported by in vitro data that suggests that in cells, where most G4s are unfolded, PhenDC3 acts as a G4 stabilizer, increasing the number of ribosomal G4s and therefore increasing the heme-binding sites on the ribosome.
Lastly, to determine the physiological relevance of the observed in vivo rRNA-heme interactions, the authors used the HS1 heme sensor to quantify bioavailable levels of cytosolic heme upon conditions in which rRNA G4s were disrupted. Exposing HEK293T cells with PhenDC3 resulted in a decrease of bioavailable heme in the cytosol, indicating rRNA G4s sequester heme and make it less bioavailable. The results also agree with existence of rRNA G4 formation in vivo. These findings have major implications, and the authors propose that heme-rRNA interactions may be important for cotranslational protein hemylation and/or for buffering cytosolic heme.
Mestre-Fos, S., Ito, C., Moore, C. M., Reddi, A. R., & Williams, L. D. (2020). Human ribosomal G-quadruplexes regulate heme bioavailability. The Journal of biological chemistry, 295(44), 14855–14865. https://doi.org/10.1074/jbc.RA120.014332
