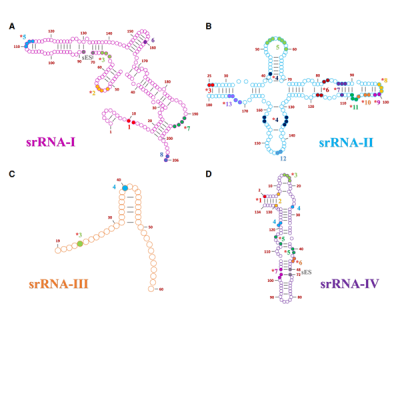
It is thought that RNA played a critical role in the origin of life but exactly how RNAs associated to form prebiotically useful RNAs is not clear. The 28S rRNA of some trypanosomatid organisms has been found to be segmented into independent rRNAs of different sizes. The larger rRNA fragments are able to join together by standard base pairing, but the smaller fragments do not appear to have these regions of interactions. Here the authors analyze the ribosomal cryo-EM structures from T. brucei, L. donovani, and T. cruzi. to identify the RNA/RNA interactions that stabilize small RNAs. Long-range interactions are a type of nonstandard interactions that might have been crucial throughout life’s history and origin. Long range RNA/RNA interactions were found to cluster into conserved regions, strengthening the smaller rRNA segments to the main scaffold. These include splayed-apart nucleotides that aid in standard and nonstandard base pairing, A-minor interactions which has been cataloged as the most abundant tertiary structure in the ribosome LSU, and standalone hydrogen bonding interactions. The most common functional group by which individual hydrogen bonds occurred was through the ribose 2’-OH which is a feature that distinguishes RNA from DNA. This is indicative of RNA’s ability to form intricate structures and complex shapes. The ribose zipper is another RNA/RNA interaction that seems to contribute to the stabilization of close contact areas. In the context of segmented ribosomes, these long-range interactions seem to converge into specific areas where two or more segments meet, favoring the stabilization of the segmented ribosome as a whole. The number of nucleotides and the nature of the interactions vary among the segments. In addition, there are differences between equivalent regions in the different organisms studied which highlight the inherent flexibility of RNA. These results help understand the versatility of RNA and the true nature of RNA sequence space.
