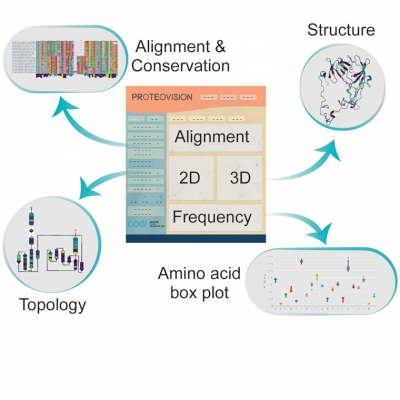
ProteoVision is a webserver that allows users to visualize proteins in primary, secondary, and tertiary forms simultaneously. These forms include multiple sequence alignments (MSA), topology diagrams, 3D representations, and amino acid frequency plots. ProteoVision links these visuals by first identifying relevant PDB structures for sequences in a selected or uploaded alignment. Next, ProteoVision searches for relevant chains within the selected PDB ID to match those in the current alignment. The server is then able to connect the selected alignment and structure by coupling the alignment indices with the residue indices from the structure. ProteoVision employs the MSAViewer which has been adapted to provide integration with other plugins via sharing data mappings, dynamic positioning, and highlighting of a specific position within the alignment. From the alignments, ProteoVision can compute phylogenetic conservation scores and physicochemical properties such as charge, hydropathy, hydrophobicity, polarity, and mutability. These properties are visualized using different color gradients in 1D, 2D, and 3D. In addition, ProteoVision can yield an interactive visualization of amino acid frequencies and their mean values for species within the multiple sequence alignment.
A principal component of ProteoVision is the DESIRE database (DatabasE for Studying and Imaging of Ribosomal Evolution) which provides a collection of ribosomal protein (rPRotein) alignments. The Taxonomy Browser can navigate rProteins across species within the database. DESIRE can link information on species taxonomy with gene annotation, polymer sequences, and ribosomal protein nomenclature. ProteoVision is not limited to rProteins; users can also upload and visualize an alignment of any protein and connect it with a 3D structure and corresponding topology diagram if available.
Users can explore protein structure and evolution and save their visualizations in a variety of formats on ProteoVision at http://proteovision.chemistry.gatech.edu/.
